On connectomes and cell lineages
Jun 10, 2011, 6:59p - Science
(If I blog rarely and if my rare posts are only about my worms, then I must simply be obsessed with them. And that's just the way it goes I guess.)
The organism, the biological being, is the most amazing object in all the world. I find it more intriguing than basic physics (which in its own right is pretty damn puzzling, esp. quantum stuff) and pretty much anything else, except for the phenomenon of consciousness.
The organism I study is called C. elegans, and I've been studying it for almost 4 years now. That seems like a long time - that's as long as undergrad and as long as my time at Google, and at both places I feel like I accomplished much more than I have so far in grad school. But I guess that's besides the point - perhaps to accomplish something truly worthwhile you must build up to that point first.
Anyhow, C. elegans is a microscopic worm that is perhaps the most tractable multicellular organism that I know of. It's the "simplest" organism as crudely measured by number of parts: 959 somatic cells of which 302 are neurons. It's the only multicellular organism for which the entire "connectome", or neural network, is known. I found that though this is true, it was very difficult to actually visualize and navigate through this network. So I wrote a simple web-app that makes browsing and querying the neural network really fast and simple. I released the app 2 years ago, and since then it's become the hub of my thinking around how the worm's brain might work, a truly indispensible tool. Of course, to the non-worm-obsessed it's probably as useful and as entertaining as foreign language refrigerator magnets.
Recently, I augmented the neural network to include not only connectivity between neurons but more information about each neuron's function. Connectivity is fine and all, but it alone doesn't tell us what each neuron is actually doing. You can uncover each neuron's function by doing experiments where you perturb specifically that neuron or it's genetic composition and observe whether anything else changes in the organism. This approach to neurobiology is about 30 years old in the history of worm science, and a lot of knowledge has been accumulated, but it's all spread across hundreds of papers that are endless streams of prose. In other words, it's difficult to quickly find information about a neuron's function, given all the text you have to wade through. If we have any hope of understanding the worm's brain in its completeness, this knowledge needs to be condensed and made more accessible to the kinds of questions we're interested in asking. In this pursuit, I've embellished my neural network web-app with functional data about each neuron, as I come across it while reading papers. For example, I recently read a paper that said if you killed neuron X, the worms would no longer respond to being poked in the back. Rather than just forget this knowledge within a few months (which is what happens to most of the knowledge I garner from papers), I now drop this little tidbit of data into the so-called "loss of function" bucket for neuron X, and then read on. In this way I've been sifting and accumulating all the structured functional knowledge which is unfortunately not available in a structured form. And as this database of neural knowledge grows, our ability to understand and predict the effects of novel stimuli and network states should also expand, without relying on storing all the relevant factoids in our heads. Though the database does not cover all known information, hopefully in a few years I will have read all the relevant papers and deposited them, such that all will be up-to-date. Then the real fun will begin.
But the neural network isn't everything - it's just the neurons. What about the remaining 600 or so cells in the worm? Good question. We know a lot about these cells as well, and our picture of the organism would be incomplete without them. To start, I've developed a visualization of the worm's cell lineage. This cell lineage, which is in the shape of a hierarchical tree, includes all the (non-germ) cells in the worm, and allows you to find out when each was born (and when each died, if appropriate) just by searching for the cell's name. I actually wrote this last summer while I was doing my starvation experiment, but was waiting to release it after I finished fixing some bugs and adding a bunch of features. Alas, my interest in developing the app has faltered, and now that it's been more than 6 months, it's timer is up and it's being released whether it likes it or not. It's in a totally usable form, but there are some rough edges which might scuff you up. Google would release it with a "beta" tag, but alas, I am not Google.
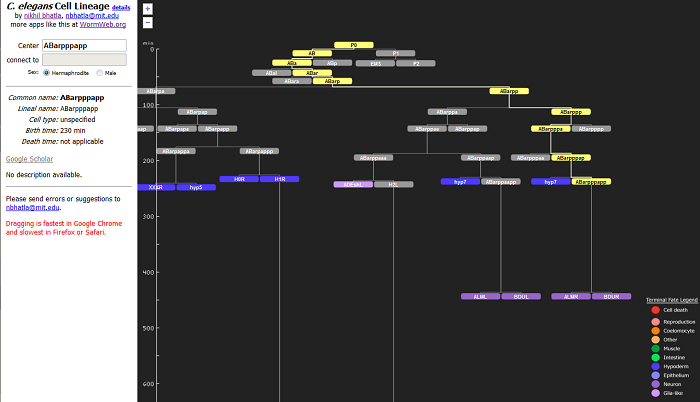
An Interactive Visualization of the C. elegans Cell Lineage
I hope all you worm researchers out there enjoy this. Gone are the days of flipping through papers to find out when that cell you're looking for is born. Remember though, you need to search for a specific cell. If you search for a cell class (e.g. ALM instead of ALML or ALMR), you're going to get nada.
For everyone else out there, click around a bit and then go on your merry way.
Enjoy. And of course comments or questions are always welcome.
--
Note: The cell lineage visualization is built on top of a customized version of the Javascript Visualization Toolkit.
Read comments (3) - Comment
Yu-li
- Jun 16, 2011, 10:23a
Hi, your post is quite interesting even though I do not fully understand it.
Here's my thought/question that just passed through my mind.
In case of Homo Sapiens(haha;;), "locked-in syndrom" happens. Even if one can no longer respond to being poked in the back or anywhere he/she still can have consciousness. (It would correspond to activities of other alive neurons though...) Motor neurons might not be an essential part of our consciousness.
So, I wonder if "locked-in syndrom" is possible for the worm. It might be hard to answer, but a necessary condition is signals in the neural network even when all the motor neurons are killed. (Life of the worm should be maintained during the experiment(?). I don't know whether it is possible.)
My thoughts are not very clear, because I do not know much about the language of neural network of the worm.
I just thought your ideas about the consciousness of worm was interesting, and I wish to read more on it. Thanks~
nikhil
- Jan 9, 2014, 6:35a
For those wondering how the data were collected: I manually analyzed the lineage diagrams from Sulston that I found in the back of Wood's C. elegans book. There was no digital resource for the lineage at the time, so I used a ruler to resolve events on the time axis down to 5 minutes.
Haoqi Chen
- May 15, 2015, 1:05a
Thank you for building this really fancy app. I'm currently analyzing the cell lineage and I wonder if your digital version of the C.elegan lineage can be adapted to statistical software like R? for example, be rewritten as a .phylo object in ape package?
« A simple web-app for counting events over time
-
A simple argument for the existence of the soul »
|