Visualizing a Worm's Neural Network
Apr 21, 2009, 11:17p - Science
For almost a year and a half, I've been working in Bob Horvitz' lab at MIT studying the nematode C. elegans. A microscopic worm of diminutive proportions (weighing in at only 1 millimeter in length), a single creature is just smaller than the size of an eyelash. These worms have been studied since the 1970s and much is known about them. They were the first multicellular organism who's genome was fully sequenced (in 1998) and their entire cell lineage is known (in other words, the pattern of cell division that takes a single fertilized egg to every cell in the worm's body). Here's a picture of an adult worm (~1000x magnification):
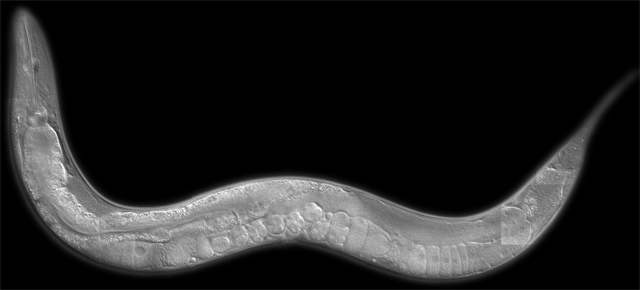
The head is in the upper left, the tail is to the right, and you can see some eggs in the mid-body.
Why do I study this worm if I'm interested in brains and consciousness? They have very few neurons (brain cells) compared to other organisms. For example, humans have billions of neurons (so many that an accurate count is difficult), while this worm has 302. That's right, each individual worm has "only" a few hundred neurons, making it a "simple" creature who's operation might one day be understood. Of course, biology is really hard, and rarely do things remain as simple as they initially seem. Nonetheless, given the fact that we know nearly nothing about the basic operations of neurons, a "simple" creature seems like a good place to start.
Because the worm has only three hundred neurons, people have been able to reconstruct how every neuron is physically connected to every other neuron (at locations called synapses). This was not a trivial task, and cumulatively took several person-decades (it was done largely by hand, without the help of computers). In studying the worm, I found that though this anatomical network is available, it isn't accessible in a form that I find easy to navigate and use. So I decided to make a web page that would be an interactive neural network. I finished the first version last night, and it's available here:
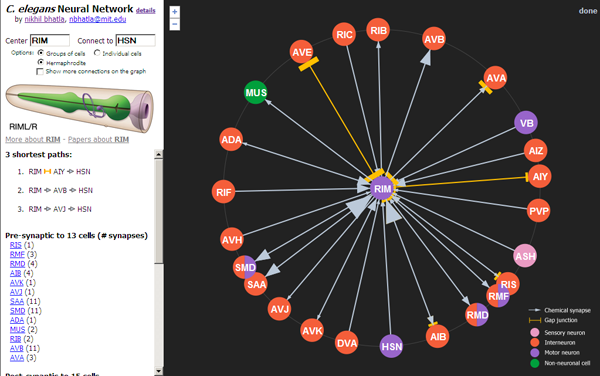
(Note that it works in Firefox, Google Chrome, and Safari, but not Internet Explorer)
Some things you can do with it:
1) Click on neurons to re-center them and see what other neurons they're connected to
2) Specify a start neuron and an end neuron to find the shortest path between 2 neurons (e.g. BAG [CO2 sensor] and HSN [egg-laying motor neuron])
3) Zoom in and out to view neurons more than just 1-degree away
4) View all connections in the visible portion of the network (makes a dense spiderweb) by checking the "Show more connections on the graph" checkbox
5) View and search the network of individual neurons. By default, neuron groups (e.g. pairs of neurons) are displayed, but this is an oversimplification, and the individual neuron network is a more accurate representation of the true network.
Notes:
- The network includes all 302 neurons and a handful of other cells (e.g. muscle).
- The network can be broken into 2 pieces: the main neural network and the pharyngeal neural network (the pharynx is the "jaws" of the worm and chews its food). These networks are thought to be connected by only a single gap junction between RIP (of the main network) and I1 (of the pharyngeal network).
- 1 neuron group (CAN) is not known to make any synapses to other neurons, so is effectively an island in the network.
- This network is only the hermaphrodite network. C. elegans also comes in a male form, and has many additional neurons not shown in this network. These may be added in the future.
- Note that the number of animals surveyed for this connectivity chart is no more than 5 and as few as 1, so what's shown here should be treated as a sampling, not a definitive network for all individuals of the species C. elegans.
- Finally, just because 2 neurons are anatomically connected does not mean that they're functionally connected, and just because 2 neurons are NOT anatomically connected does not mean that they are NOT functionally connected.
Known issues:
- Re-centering the network can get really slow if you click on individual neurons in the pharynx (e.g. MCL). Be patient as it may take several minutes for your click to be processed.
- You can only search for paths between neurons that can be found by the program within 60 seconds, which is a small subset of all possible paths. Sometimes it may timeout, yielding no paths if the neurons are sufficiently far away from each other.
- The data set isn't 100% accurate. I've recently noticed that some of the Chklovskii data (see below under technical details) is missing some of the data from the White 1986 paper (also below), data that shouldn't be excluded based on the criteria they provide. I'm looking into this.
I hope you find this interactive neural network interesting, thought-provoking, and, if you're a C. elegans researcher, maybe even useful.
----
Technical details:
- Complete list of cell types included in the network:
- 302 individual neurons in 118 classes
- 7 pharyngeal muscles
- 2 pharyngeal marginal cells
- 2 pharyngeal gland cells
- 1 pharyngeal basement membrane
- 1 muscle class
- Data sources: I relied on 2 sources for the data displayed in this visualization:
1) Main network data is from the Chklovskii group and was published in Chen..Chklovskii 2006. I modified the raw data file a bit to ease the processing of it, and it is available here. Details for how to read the file are here. A list of neurons and the associated classes is here, modified from the Japanese CCEP group. Note that this data file includes the work of White..Brenner 1986, Hall & Russell 1991, Hobert & Hall 1999, Durbin 1986, and Achacoso & Yamamoto 1992.
2) Pharynx network data is from Albertson & Thomson 1976. My data file is here, which was modified from the Japanese CCEP group. This data is older and much less clean than the first data set. Some of the issues are:
a) In Albertson 1976, some synapses are recorded as being between a single cell and a cell class. In these cases, connections are assumed to be to all cells of that class (the most conservative assumption given the data, I think).
b) Sometimes the type of synapse is missing. In these cases, I either default to a pre-synapse, or use the comment made in the text (but missing in the figure).
c) A synapse is annotated as both a gap junction and a chemical synapse. In these cases, I assume both are present. This causes double-counting of some synapses, so the synapse count for the pharynx isn't very accurate.
d) Often the target muscle name is missing. In these cases, I direct the synapse to the muscle that is inferred by the figure. Also, sometimes the annotations were wrong from CCEP, so I fixed them.
e) I renamed several of the muscles and other non-neuronal cells to names that don't collide with and are more consistent with existing neurons.
f) I count each line of the data file as a single synapse
g) Note that many synapses in the pharynx are highly variable from animal to animal (i.e. sometimes they're there and sometimes they aren't).
- Data visualization: For the fancy animation and radial graph, I built off of an existing javascript library called JIT. I modified only a small part of it to accommodate my use, and greatly appreciate the fact that this library was available, open source, and free to use.
Read comments (17) - Comment
Sundar
- Apr 22, 2009, 6:31p
Very cool, nice to see you doing great work. thanks for the chrome plug:)
Ruggero
- May 15, 2009, 12:46a
Simply fantastic.
Matej
- Sep 5, 2009, 4:33p
I'm involved in neural networks in computing (artificial NN in chips) and this is very interesting for me.. Nice!
Carlos
- Nov 30, 2009, 4:10a
Excellent iniciative!
Diablo
- Sep 5, 2010, 2:13p
Interesting.
Drew Barfield
- Oct 6, 2010, 2:12p
Awesome. A very useful tool.
Gokul Rajan
- Nov 16, 2010, 7:59p
This is simply fantastic! Now i know how your engineering skills are helping you here! :) great work!
Jason Toy
- Jan 30, 2012, 11:00p
great work
jaronimoe
- Jul 19, 2012, 12:35p
Hey nice work!
Did you by any chance publish an article about this tool of yours at a conference?
I would rather cite an article than a webpage ;)
nikhil
- Jul 21, 2012, 9:31p
Nope, no article yet. It's still a work in progress, and there are a few more features I want to add. I'll post a link here if I publish it in the old-school way.
What's wrong with citing a webpage?
jaronimoe
- Jul 24, 2012, 5:30a
There's nothing wrong with citing a webpage but my university prefers literature citations since articles are reviewed and webpages "grow in the wild" - e.g. anybody could state anything on a webpage.
But since I'm citing a visualization application and not some scientific statements it should be no problem.
Thanks again!
jaronimoe
- Jul 24, 2012, 5:36a
I forgot to ask:
what does the size of the arrow tip represent?
nikhil
- Jul 26, 2012, 8:26a
The size of the arrow is proportional to the number of chemical synapses between two neurons.
Likewise, the size of the orange bar is proportional to the number of gap junctions between the two neurons.
Often we assume that the more synapses that exist between a pair of neurons, the more likely the neurons are functionally connected. But this isn't necessarily the case, just a place to start thinking.
Harry C. Pedersen
- May 7, 2015, 5:09p
Good job! But why such a complicated organism? Why not some thing with only a single neuron? Yes, I know , it is not a "net", but, it does interact, with the environment, in some simple way. And it interacts, with the parent organism, in some way. Hopefully a simple to understand way. The knowledge thus gained, may just pave the way, to a little more complicated network, one, perhaps with 2 neurons. Thus, at last, we come to a neural nat work.But, with some use full knowledge. And maybe, if we are lucky, some sensible rules. This, is, of course, just a suggestion. I am not an expert, or any thing , but, may i suggest a possible starting point? The single celled organism, known as the "euglena
". these things operate as plants, when light is present, but act as animals, when light, is absent.How this happens, is not as simple, as it at first appears. Yes , the chlorophyl molecule, can act as a switch. But, what exactly gets switched? I asked this question , in high school biology class, and got booted out, as a "wise guy. But I guess my timing was off, as this was 1957, and I have to assume, that no one was quite ready to answer such questions, back then. At any rate, thanks for hearing me out. Harry
Nic
- Sep 9, 2021, 2:23a
Is this model now basically complete, in terms of the basic connectome?
nikhil
- Sep 9, 2021, 7:50a
The model is complete. However, as researchers generate and/or analyze new datasets, new and different connections are being found. See Cook..Emoons 2019. Whole-animal connectomes of both Caenorhabditis elegans sexes.
Available at https://www.nature.com/articles/s41586-019-1352-7
Doublestar Rock
- Jun 2, 2023, 10:42a
very interesting work!
« You have 2 time travels: Choose wisely
-
To Be Conscious in a Body, Frozen »
|